
#Palindromic sequence how to
NOTE:- Time Complexity of the above implementation is O(n^2) which is much better than the worst case time complexity of Naive Recursive implementation.Count the total number of Palindromic Subsequences: Here, we are going to learn how to find the total number of Palindromic sub-sequences in a string? This is a standard interview problem that can be featured in any interview coding rounds. OUTPUT:- The length of the Longest Palindrome Subsequence is 5. On the other hand, sequencing-by-synthesis sequencers had no difficulty in doing so. * *Driver program to test above functions* */Ĭout << "The length of the Longest Palindrome Subsequence is " << longestpalindrome(arr)) Palindromic sequence impedes sequencing-by-ligation mechanism Taken together, we demonstrate that SOLiD machines, which employ sequencing-by-ligation mechanism, are unable to read through the palindromic region. manner similar to Matrix Chain Multiplication DP solution useless and not filled in the process. Note that the lower diagonal values of table are Choice A shows the WT with the sequence, but the mutant stand without it. This question also requires knowledge in what palindromic sequences are used for - in this case determining if a gene is mutated via southern blot. Strings of length 1 are palindrome of lentgh 1 The presence of a palindromic sequence is only part of the story. Int L // Create a table to store results of subproblems coli chromosome DNA, accounting for about 1 of the genome. There are approximately 5001000 copies of REP sequences interspersed on E. The REP sequence is about 40 base pairs long. Returns the length of the longest palindromic subsequence in seq Repetitive extragenic palindromic (REP) DNA sequences were firstly identified in Escherichia coli and Salmonella typhimurium 65. Certain sequences of DNA are the same whether they are read 5' (five-prime) to 3' (three prime) or 3' to 5'.

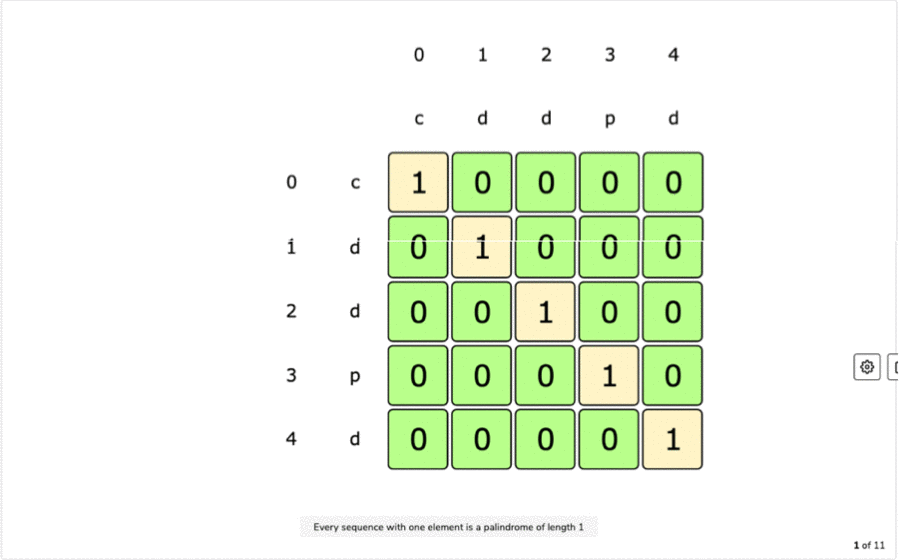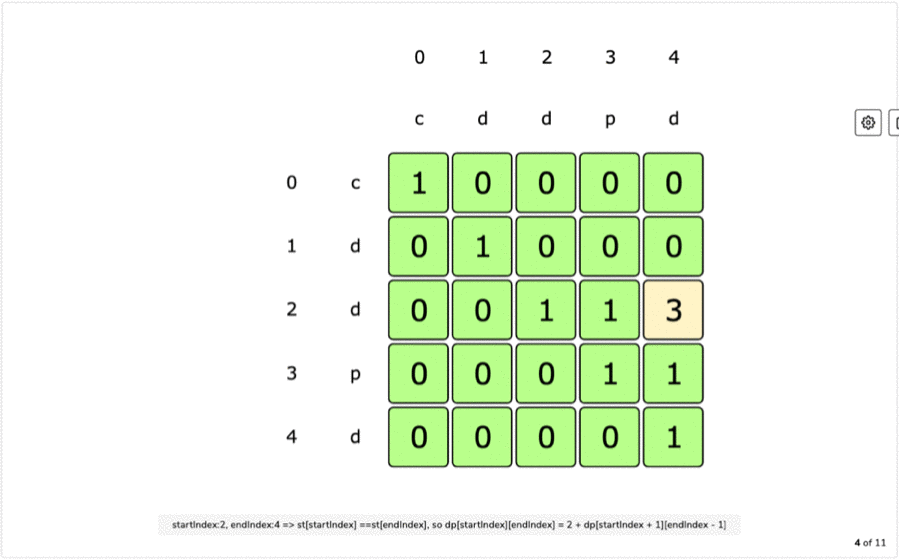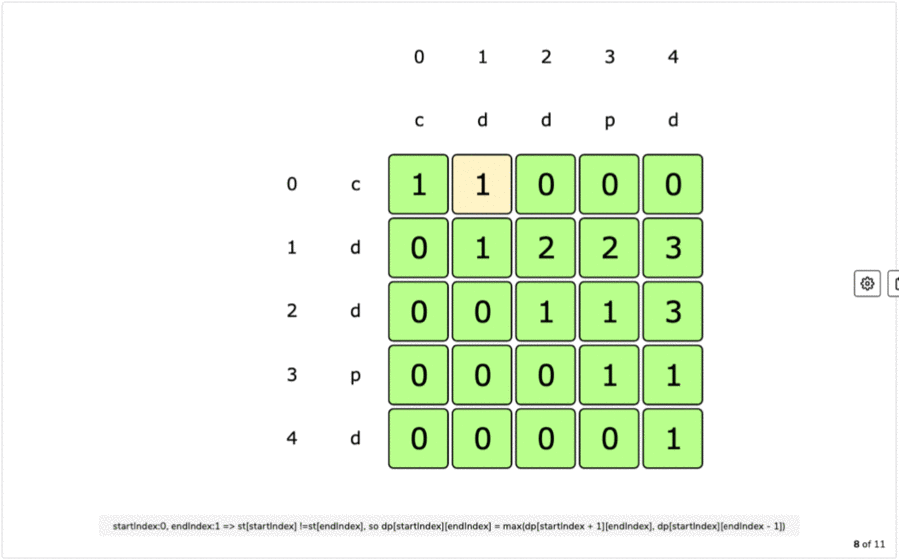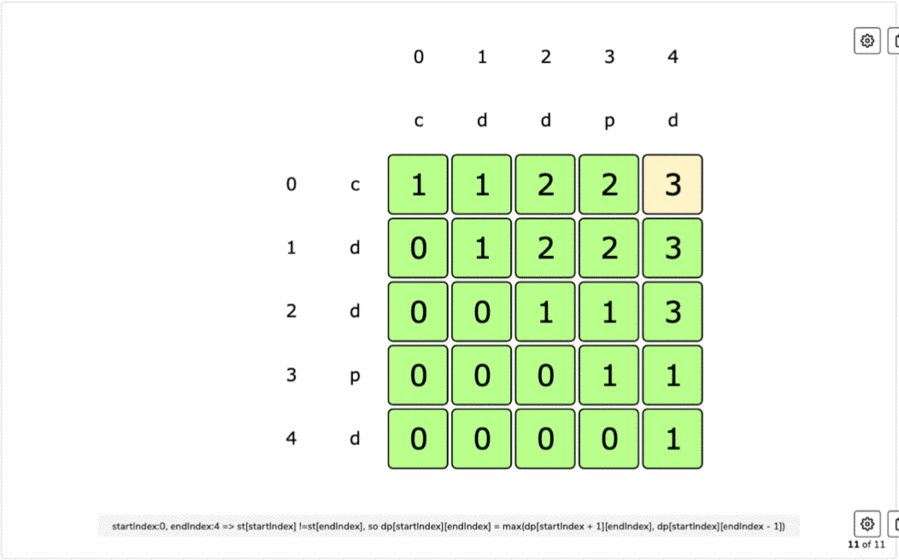
IF first and last characters are not same a palindrome is a word that is the same forward as it is backward such as the word racecar, the name Otto, or the saying attributed to Napoleon 'Able was I ere I saw Elba'.L(i, i) = 1 for all indexes i in given sequence Every single character is a palindrome of length 1.removing the first character and recursing for the remaining substring X.removing the last character and recursing for the remaining substring X.If last character of string is different from the first character, we return maximum of the two values we get by: For example, restriction enzymes often recognize palindromic sequences of DNA. They occur in genomes of all organisms and have various functions. If last and first character of string is same, we can include both characters in palindrome and recurse for remaining substring X Palindromes in DNA consist of nucleotides sequences that read the same from the 5'-end to the 3'-end, and its double helix is related by twofold axis. The idea is to compare the last and first character of the string. Here, we can use Recursive method to solve this problem. This solution is exponential in term of time complexity. Due to the palindromic sequences often found on ChrY, Y-STR loci can sometimes exhibit as. The naive solution for this problem is to generate all subsequences of the given sequence and find the longest palindromic subsequence. A palindromic sequence of 146 nt is at the 3-end of the genome and a different palindromic sequence of. The longest palindromic subsequence is BABCBAB. The length of longest palindromic subsequence is 7. The longest palindromic subsequence is BCACB. The length of longest palindromic subsequence is 5. the other is a dynamic programming approach where we will reduce the time complexity from O(2 N) to O(N 2).If last and first characters of X are same, then L(0, n-1) L(1, n-2) + 2. Unlike substrings, subsequences are not required to occupy consecutive positions within the original sequences. Let X0.n-1 be the input sequence of length n and L(0, n-1) be the length of the longest palindromic subsequence of X0.n-1. This can be done using Dynamic Programming in O(N 2) time. In a single-stranded molecule, COMPLEMENTARY BASE PAIRING can occur when the chain is folded back (See also HAIRPIN ). The T protein binds this sequence as a dimer, with each monomer binding a half site called a T-half site (5’-AGGTGTGAAATT-3’) ( Kispert and Herrmann, 1993 ). The longest palindrome subsequence problem (LPS) is the problem to find the length of longest subsequence in a string that is a palindrome. palindrome a sequence in double-stranded nucleic acids that reads the same on both strands when reading one strand from left to right and the other from right to left (i.e. A palindromic DNA consensus sequence with high affinity for T protein was first defined and called the T-box binding element (TBE). Reading time: 20 minutes | Coding time: 10 minutes


 0 kommentar(er)
0 kommentar(er)
